DrD, your R code does not work
Short response: Code snippets from preformatted text will copy and paste from your computer’s clipboard and run correctly in R without need for further editing.
Longer response
Every effort has been made to provide you with verified code. While it is not great practice to copy and paste code to run, we all do it so that we can start to learn how to code (and how to troubleshoot!). However, when a code snippet copied from my site fails to run as predicted, you shouldn’t first think that “DrD made a mistake.” The error is most likely from the copy/paste result.
As you know, R expects text only and will fail to run script, returning an error message, if anything but text is present. Before you notify me about “errors with your code,” first check that your copy and paste effort has not introduced incompatible formatting. It’s really the first thing you should think about while troubleshooting — is there a formatting problem, did non-text characters creep into my R script?
I use WordPress with my sites. Between WordPress conventions and the theme I use, some modifications are done by me to make the code present correctly on the webpage. Common copy/paste errors with R code include
- Tables on the web may not copy/paste correctly into your script file.
- Standard quotes replaced by smart quotes (see Wikipedia)
While there are some advanced tricks you can do (e.g., view web source), you can tell by looking at the formatting whether or not you’ll be able to do a clean copy and paste from your clipboard, or whether you’ll need to edit to clean up the code. Look for preformatted block of text, which presents the text exactly as intended. The block appears as gray box and the text, preformatted, looks like it came from an old typewriter.
I’ll show you an example. The first version of the code presentation, “Web formatted version,” will fail to run in R unless you edit the defects. The second, “Preformatted version,” will run without error.
Consider the following small data set, Carbon Dioxide production (microliters per minute) at room temperature (22 °C) by 0.5 g yeast (Saccharomyces cerevisiae) metabolizing two different carbon sources, table sugar and Stevia in the raw (which actually contains a small amount of glucose!). Two versions of R script to read the data in table form and save to object myData are provided below.
Web formatted version
myData <- read.table(header=TRUE, sep=”\”, text=”
| Minutes | Sugar | Stevia |
| 0 | 0.0000 | 0.0000 |
| 30 | 16.6667 | 6.6667 |
| 60 | 12.5000 | 5.8333 |
| 90 | 5.5556 | 2.2222 |
| 120 | 6.2500 | 3.3333 |
| 150 | 3.6667 | 1.3333 |
“)
Copy and paste R code snippets from the Web formatted version will fail to run in R unless you note and correct the problems with formatting. See screenshot of R script from copy/paste of Web formatted version (Fig. 1).
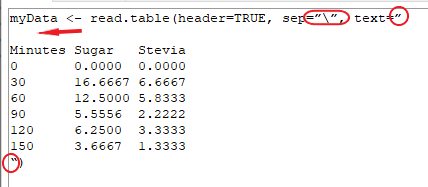
Figure 1. Failed R code snippet copy paste to script window. Circles highlight smart quotes; arrow points to blank row inserted by paste.
Preformatted version
myData <- read.table(header=TRUE, sep="\t", text=" Minutes Sugar Stevia 0 0.0000 0.0000 30 16.6667 6.6667 60 12.5000 5.8333 90 5.5556 2.2222 120 6.2500 3.3333 150 3.6667 1.3333 ")
Copy and paste R code snippets from the Preformatted version will run in R without need for you to edit. See screenshot of R script from copy/paste of Preformatted version (Fig. 2).
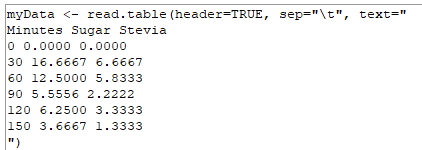
Figure 2. Successful R code snippet copy paste to script window. Note straight quotes and continuous script of code.
/MD